Introduction To High Content Imaging
Introduction
High content imaging (HCI) is an area of imaging where the aim is to maximize data capture. Any kind of imaging can be high-content if the objective is to obtain as much data as feasibly possible, regardless of imaging system, sample, magnification, fluorophores, and camera used. This makes specifics in HCI difficult to define, but in general, HCI involves performing normal imaging thousands or millions of times in order to maximize data capture effectively.
An analogy for HCI is to image a football field with dinner plates laid across the whole field. Researchers doing standard imaging and analysis may only be able to look at a few plates, but HCI allows the whole field to be measured. The whole field could be a single cell culture dish (where each plate is a cell), or the whole field could be a multi-well plate (where each plate is a well/experiment).
Where a standard live-cell experiment may observe a small number of cells dividing, in HCI the aim is to image hundreds of cells dividing in order to best observe how they divide. This makes HCI most useful for high throughput applications in order to maximize the number of options for better statistical analysis. Examples include drug discovery, where many variations of chemicals can be imaged; and imaging dense tissues at high magnifications, where many images are needed to stitch together the large sample; or lab-on-a-chip imaging, where many experimental variables can be altered across many experiments.
HCI is often used with cell biology experiments in combination with automation and quantitative data analysis. The automation can come from the hardware, such as with automated motorized microscope stages, or from the software, with automated image recognition via machine learning for more high-throughput analysis. The efficiency that allows for high levels of data capture can be gained by introducing automation at various stages of the imaging process.
HCI Example
An example of HCI can be seen in Fig.1 from Carty et al. (2015), where a specific type of neuron (medium spiny neuron) was identified, characterized, and quantified with HCI of an entire mouse brain slice (Fig.1A). This study involved:
- Slicing a mouse brain and culturing the slices in neural growth medium
- Staining each slice with multiple fluorescent markers for specific proteins
- Acquiring hundreds of images at different XY positions at 40x magnification
- Stitching these images together to get an overall image of an entire brain slice
- Performing quantitative image analysis on specific fields with specialized software
- Determining cell number, sub-cellular localization, cell shape (morphology), cell size, locations of co-staining, presence/absence of certain cell types, clustering in certain brain regions, etc.
- Development of a customized automated script analysis to identify desired regions
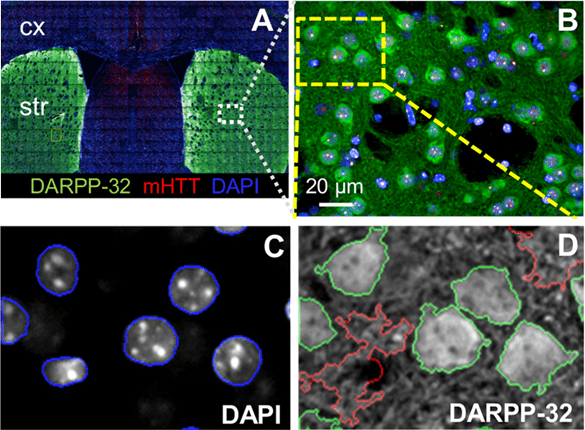
Figure 1: An example of HCI on the brain of a zQ175 mouse. A) Brain slices were stained with DAPI (nucleus marker),
DARPP-32 (striatal neuronal marker), and mHTT (Huntington's disease marker), localizing to the cx (cortex) and
str (striatum). The entire slice was imaged at 40x with multi-field imaging and then stitched together (as seen by image borders).
B) A single 40x field of view from the slice seen in A, showing individual cells and networks. C) A selection of cells from B,
showing the location of the cell nucleus via DAPI staining. D) Neurons identified by the intensity of DARPP-32 staining.
Image adapted from Carty et al. (2015).
HCI is a good fit for imaging brain slices, which can often be extremely dense with cells and structural/functional relationships. In the case of Fig.1, HCI allowed for automated multicolor, multi‑parameter fluorescence imaging, followed by a quantitative analysis of thousands of cells in multiple brain sections from multiple animals. This HCI approach is efficient and thorough when working with this volume of data as some alternative approaches would take far longer.
This HCI example shows that quantitative data on a range of functional and morphological changes in cells can be obtained through the collection and analysis of many images, often in real-time. This involves standard fluorescence detection, but also automated microscopy and multi-parameter algorithms in order to process large datasets. This makes the software used in HCI important, which is covered in more detail in our separate article on analysis programs for HCI. An idea of the workflow involved in HCI can be seen in Fig.2.
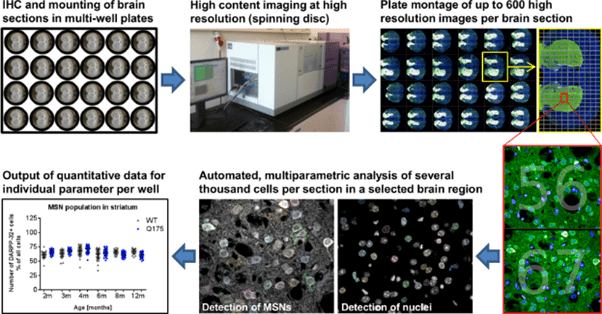
Figure 2: A workflow for HCI. IHC: immunohistochemistry. Slices of mouse brain were stained and mounted in a
glass-bottom multi-well plate, and then automatically imaged at high resolution with a confocal spinning disk system.
Up to 600 images are taken per brain section, often high magnification images stitched together. These high
magnification fields of view are analyzed, with software studying several thousand cells at a time via automated,
multi-parameter software analysis on every single image. The comprehensive quantitative data sets were
gathered for numbers, subcellular structures, morphology, and intensities for stained subpopulations of cells per
slice per well. Image from Carty et al.(2015).
Drug Discovery
HCI is useful at the early stages of drug discovery, giving useful direction for future experiments by using high-throughput image analysis of thousands of different compounds on cells to brute force solutions.
By treating cells with different pharmaceuticals and imaging the effects on the cell phenotype; or by labeling specific disease marker proteins and imaging the effect of different chemicals, drug discovery can be accelerated in the pre-clinical phase. HCI could measure cell number, protein aggregates, nuclear morphology, and toxicity measurements. Identifying drug targets or mechanisms of action early on accelerates the entire process. This is a vital application, as the current drug discovery model can take tens of years in order to develop better solutions for certain diseases that can result in a much lower quality of life.
Alongside drug discovery, some of the main uses of HCI are studying cell behavior, mechanisms/molecular function, and safety/toxicology studies. These studies are done with a number of fluorescent markers with an aim to study anywhere from 3‑20 different parameters.
Issues with HCI
Due to the large numbers of samples and images, performing HCI can be time-consuming, requires a large volume of digital storage, and requires specialized software in order to automatically analyze data with multiple parameters. While the acquisition of images with a fast camera and imaging system can be streamlined and efficient, there is a bottleneck with analysis and data transfer, storage, and handling.
Summary
High content imaging aims to maximize data capture within imaging experiments. This can be done temporally by maximizing imaging efficiency, minimizing dead time, and performing normal imaging experiments thousands or millions of times in order to gather a large enough effective sample size. High content imaging allows for high throughput experiments, useful in certain applications such as neuroscience, drug discovery, and lab-on-a-chip experiments with many variables. See the other articles in our High Content Imaging sections to learn about hardware, analysis programs, and suitable cameras for this format of imaging.
References
Carty N, Berson N, Tillack K, Thiede C, Scholz D, Kottig K, Sedaghat Y, Gabrysiak C, Yohrling G, Kammer Hvd, Ebneth A, Mack V, Munoz-Sanjuan I and Kwak S (2015) Characterization of HTT Inclusion Size, Location, and Timing in the zQ175 Mouse Model of Huntington´s Disease: An In Vivo High-Content Imaging Study. PLOS ONE 10(4): e0123527. https://doi.org/10.1371/journal.pone.0123527
Further Reading
Back To High Content Imaging
Join Knowledge and Learning Hub