Single Molecule FRET
Dr. Ehud Isacoff
Director, Helen Wills Neuroscience Institute, UC Berkeley
Background
Prof. Ehud Isacoff's lab at the University of California, Berkeley, USA is interested in the mechanistic properties of metabotropic and ionotropic receptor subunits and the role the resultant protein complexes play in signal transduction. His group uses a combination of microscopy approaches including single molecule Föester resonance energy transfer (smFRET) in live cells, and in vivo imaging in a variety of species. They've developed a number of tools for the investigation of neuronal function. Having pioneered the manipulation of receptors with tethered ligands, more recently they've developed photoactivatable versions of sensors such as GCaMPs.
The effects of subunit composition on metabotropic receptor transduction can be subtle and impossible to separate in mixed populations of receptor subunits. In the brain of mammals, metabotropic glutamate receptor subunits have mixed and overlapping expression patterns, making it very difficult to understand the roles of particular subunits. Single molecule in vitro analysis of receptors with known subunit compositions is needed. This allows the full characterization of biophysical properties, such as conformation change kinetics, which can be measured by smFRET.
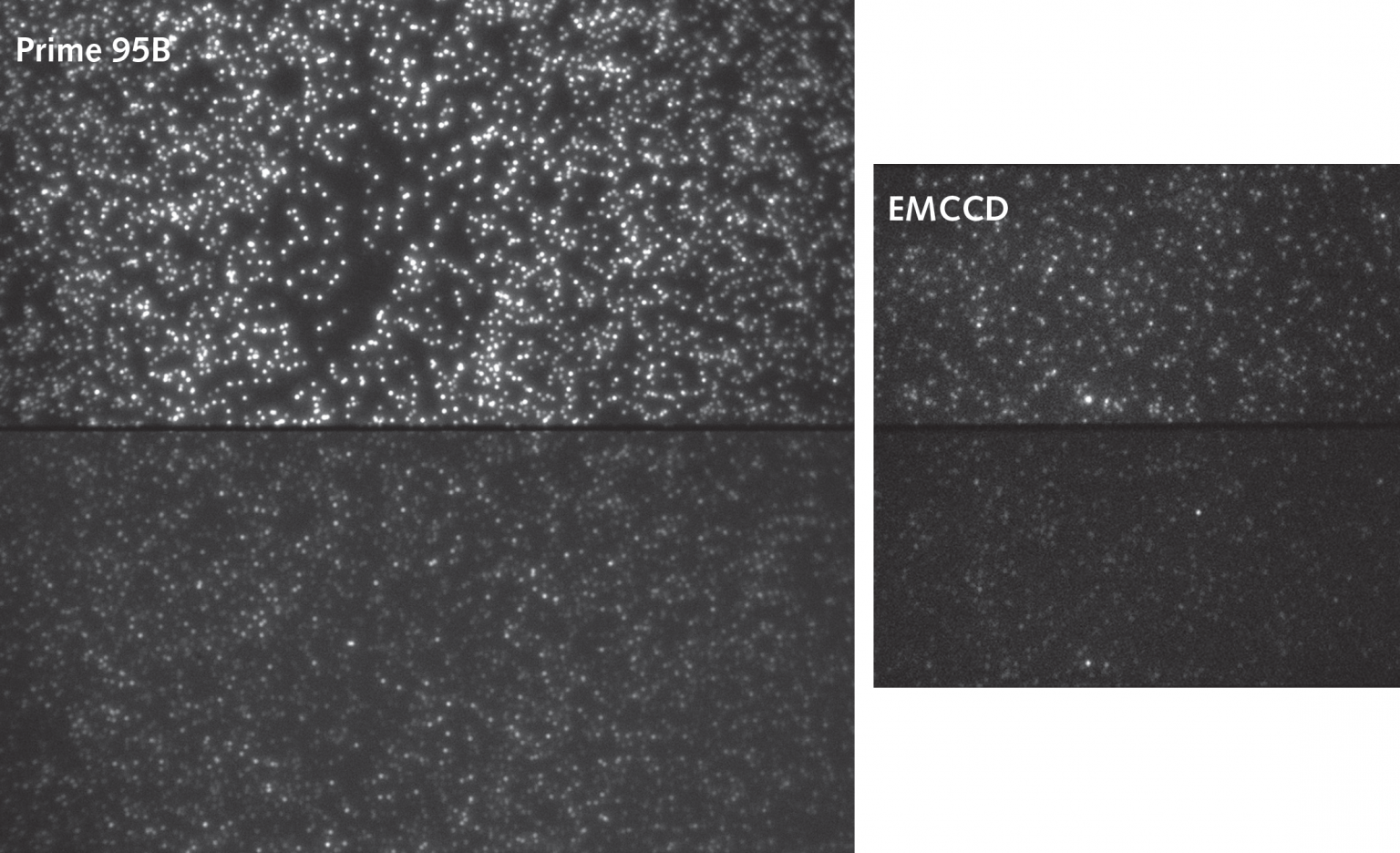
Figure 1: Donor and FRET signals separated to the top and bottom halves of the camera sensor. Comparison images were acquired on the Prime 95B sCMOS (left) and a 512×512 EMCCD (right) using the same exposure time. The increased field of view is obvious and subsequent image analysis revealed that the Prime 95B also had a higher signal-to-noise ratio. The Prime 95B is also capable of far higher speeds. This allows for higher quality data to be acquired and more protein complexes to be measured with faster sampling kinetics.
Challenge
To measure the conformation changes of single receptors, fluorescent receptor subunits expressed in bacteria are reconstituted into functional receptor types and immobilized on a surface for imaging. Specific manipulations like ligand applications can be performed to stimulate the receptors to change conformation, which is recorded in the FRET signal. The kinetics of the conformational change are accurately mapped by the temporal FRET response.
Single molecule FRET presents very specific problems for imaging. There are very few fluorophores present in each protein complex. Secondly, receptor kinetics can be extremely fast. High camera frame rates are needed to accurately sample fast events. This means exposure times are short and detecting the few photons being emitted is difficult. EMCCD cameras have traditionally been used due to their sensitivity, but they lack the speed to detect very fast events.
We have been using 10 ms exposure and getting 100 fps and I’m just as confident in the data at 100 fps with the Prime 95B as with 10 fps with the EMCCD.
Dr. Ehud Isacoff
Solution
Using the Prime 95B back-illuminated sCMOS camera, the Isacoff lab improved the speed at which they can record in smFRET experiments. Previously they achieved 10 fps with an EMCCD. They shared, "We have been using 10 ms exposure and getting 100 fps and I'm just as confident in the data at 100 fps with the Prime 95B as with 10 fps with the EMCCD." This increase in frame rate, whilst still maintaining the high signal-to-noise ratio needed to reliably record FRET changes, has allowed detection of events of 10 fold shorter duration than before.
This has allowed the group to confirm the kinetics of slower moving protein complexes, but also determine faster kinetics of other complexes for the first time. The combination of large 11µm pixels with high 95% quantum efficiency and fast frame rates make the Prime 95B an excellent choice for smFRET where both sensitivity and speed are necessary to produce high-quality data.

Learn More About the Prime 95B
Download This Customer Story