Characterization of Soil-Grown Root Systems
A Quantitative Comparison of Different Scientific CCD Types
Overview
Recently, a team of scientists working in North America and Europe developed an innovative growth and imaging platform, known as GLO-Roots, that allows root architecture and gene expression to be studied in soil-grown plants.
GLO-Roots (Growth and Luminescence Observatory for Roots; U.S. patent application: 13/970,960) is a collaborative effort between the labs of Dr. José Dinneny at the Carnegie Institution for Science (Stanford, California), Dr. Rubén Réllan-Álvarez (Langebio, Mexico), and Dr. Guillaume Lobet at the Université de Liège in Belgium. Designed and built with help from BioImaging Solutions, Inc. (San Diego, California), the researchers' new growth and imaging platform supports several model organisms, namely, Arabidopsis thaliana, Brachypodium distachyon, Setaria viridis, and Lycopersicon esculentum.1
GLO-Roots employs luminescence-based reporters and a pair of Princeton Instruments back-illuminated CCD cameras to enable studies of root architecture and gene expression patterns in soil-grown, light-shielded roots. Custom-designed image analysis algorithms allow the spatial integration of soil properties, gene expression, and root system architecture traits. The GLO-Roots developers assert that the new platform can offer great utility for presenting environmental stimuli to roots in ways that evoke natural adaptive responses and in providing tools for investigating the multidimensional nature of such processes.2
This application note will outline some of the research conducted by the GLO-Roots team, which is led by Dr. José Dinneny at the Carnegie Institution for Science's Department of Plant Biology on the Stanford University campus
GLO-Roots Setup
The GLO-Roots platform comprises four parts: (1) growth vessels, called rhizotrons, that permit plant growth in soil as well as imaging of roots; >(2) luminescence-based reporters that allow various aspects of root biology to be tracked in living plants; (3) GLO1, a luminescence-imaging system designed to automatically image the rhizotrons; and (4) GLO-RIA, an image
analysis suite designed to quantify the root systems imaged utilizing GLO1.2 An example of an experimental setup using the GLO-Roots platform is presented in Figure 1.

Figure 1. GLO-Roots growth and imaging system. (A) Three-dimensional
representation of the different physical components of the rhizotron,
including plastic covers, polycarbonate sheets, spacers, and rubber U-
channels. (B) A 35-day-old plant in rhizotron with black covers removed.
(C) Top view of holding box with 11 rhizotrons. (D) In-vivo emission
spectra of different luciferases used in this study. Transgenic homozygous
lines expressing the indicated transgenes were grown on agar media for
8 days. Luciferin (300 μM) was sprayed on the seedlings; plates were
kept in the dark and then imaged for 2 sec at wavelengths ranging from
500 to 700 nm.Five intensity values were taken from different parts of the
roots of different seedlings and averaged.Relative maximum intensity
values are indicated in lower-right graph. (E) GLO1 imaging system
composed of two Princeton Instruments PIXIS:2048 back-
illuminated CCD cameras (a) cooled down to -55°C. A filter wheel
(b) allows spectral separation of the different luciferases. On the right, a
rhizotron holder (c) is employed to position the rhizotrons in front of the
cameras. A stepper motor (d) rotates the rhizotron 180° to image both
sides. (F) A 21 DAS (i.e., 21 days after sowing) plant expressing
reporter ProUBQ10:LUC2o was imaged on each of two sides of the
rhizotron; luminescence signal is colorized in green or magenta to
indicate side.In the middle of the panel, a combined image of the two
sides is shown. The inset shows a magnified part of the root system.
Diagrams, photos, and data courtesy of Dr. José Dinneny, Carnegie
Institution for Science. First published in R. Rellán-Álvarez et al. 2015.
"GLORoots: an imaging platform enabling multidimensional characterization
of soil-grown root systems." eLife 4 (1): e07597.
Whereas most commercially available luminescence imaging systems for biomedical research are optimized for imaging horizontally held specimens or samples in microtiter plates, placing the rhizotrons in this position would induce a gravitropic response in plants. Therefore, GLO-Roots utilizes a custom-designed imaging system (known as the Growth and Luminescence Observatory 1, or simply the GLO1) that has been optimized specifically for imaging dual-reporter luciferase expression in the GLO-Roots rhizotrons.2
Two PIXIS:2048 back-illuminated CCD cameras from Princeton Instruments are mounted on top of each other so as to capture partially overlapping images of a rhizotron, while a motorized stage automatically rotates the rhizotron to capture images of both sides (refer to Figure 1E). A composite image is then generated from the images captured of each side. Figure 1F shows that approximately half of the root system is revealed on each side, with a few roots being visible on both sides. The soil sheet is thick enough to block light from portions of the root system, yet thin enough to ensure its continuous structure can be compiled from opposite-face views. The entire GLO1 imaging system is enclosed in a light-tight black box that is equipped with a door to permit the loading and unloading of rhizotrons.2
Not only do the PIXIS:2048B cameras afford the GLO-Roots researchers the desired spatial resolution and field of view, but also the excellent low-light-level sensitivity required to detect the emission wavelengths of the different reporters used. Table 1 lists the luciferases employed for the Arabadopsis-related GLO-Roots study.
| Luciferase | Origin | Maximum Wavelength (nm) | Substrate |
| PpyRE8 | Firefly | 618 | D-luciferin |
| CBGRed | Click Beetle | 615 | D-luciferin |
| Venus-LUC2 | FP + Firefly | 580 | D-luciferin |
| LUC (+) | Fireflu | 578 | D-luciferin |
| CBG99 | Click Beetle | 537 | D-luciferin |
| Lux operon | A. fischeri | 490 | Biosynthesis pathway encoded with operon |
| NanoLUC | Deep sea shrimp | 470 | Furimazine |
Data and Results
Root systems develop different root types, each of which individually senses myriad local environmental cues and integrates its environment's information with systemic signals. This highly complex multidimensional amalgam of inputs enables continuous adjustment of root growth rates, direction, and metabolic activity that define a dynamic physical network.2
Figure 2 presents time-lapse imaging of Arabidopsis root systems captured with the GLO1 system along with quantification performed using the GLO-RIA image analysis suite.

Figure 2. Time-lapse imaging of Arabidopsis root systems and quantification using
GLO-RIA. (A) Typical daily time-lapse image series from 11 to 35 DAS of a
ProUBQ10:LUC2o Col-0 plant. (B) Average directionality of three root systems
imaged in time series as in panel A calculated using the directionality plug-in
implemented in GLO-RIA. (C) Color-coded projection of root growth using the
images in panel A. (D) Root system depth, width, and area are automatically
calculated from the convex hull, which is semi-automatically determined with
GLO-RIA (n = 3). Primary root length, lateral root number, and number of lateral
roots divided by the primary root length were quantified manually.
A local polynomial regression fitting with 95% confidence interval
(gray) was used to represent the directionality distribution curve. 0° is the
direction of the gravity vector.
Diagrams, photos, and data courtesy of Dr. José Dinneny, Carnegie Institution
for Science. First published in R. Rellán-Álvarez et al. 2015. "GLORoots: an
imaging platform enabling multidimensional characterization of soil-grown root
systems." eLife 4 (1): e07597.
Here, root traits such as directionality can be observed through later stages of plant development (notice the 35 DAS root system image and the 35 DAS directionality analysis shown in Figures 2A and 2B, respectively). A time series ning 11 to 21 days after sowing Arabadopsis accession Col-0 roots expressing reporter ProUBQ10:LUC2o is presented in Figure 2A and Figure 3; a color-coded time projection is shown in Figure 2C.
Directionality analysis indicates a progressive change in root system angles, from 0° (vertical) to 55°, as lateral roots take over as the predominant root type. Figure 2D shows the evolution over time of several root traits that can be automatically captured by GLO-RIA (depth, width, area) and others that were manually quantified (primary root growth rate or number of lateral roots per primary root.2
To date, the GLO-Roots team has published data pertaining to the continuous imaging of root growth; the root system architecture of different Arabidopsis accessions; the utilization of spectrally distinct luciferases to capture additional information associated with gene expression patterns, characterization of root system interactions, and microbial colonization; adaptive changes in root system architecture under water deficit, phosphorus deficiency, and light; the suitability of the GLO-Roots platform for studying additional plant species; and more. The research team is also highly interested in discovering how other environmental stimuli affect root growth and whether such responses differ between accessions of Arabidopsis.2
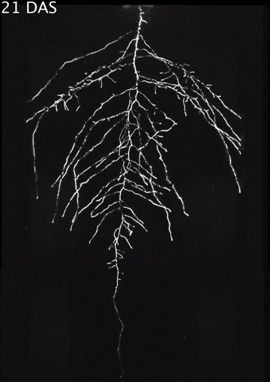
Figure 3: Time stamp from video time lapse from
11 to 21 DAS of a Col-0 plant expressing ProUBQ10:
LUC2o grown in control conditions. Video courtesy of
Dr. José Dinneny, Carnegie Institution for Science
. First published in R. Rellán-Álvarez et al. 2015.
"GLO-Roots: an imaging platform enabling
multidimensional characterization of soil-grown
root systems." eLife 4 (1): e07597.
For more data and an in-depth discussion of results, please refer to R. Rellán-Álvarez et al. 2015. "GLO-Roots: an imaging platform enabling multidimensional characterization of soil-grown root systems." eLife 4 (1): e07597.
Enabling Technology
Each of the PIXIS:2048 cameras integrated in the GLO1 imaging system utilizes a CCD featuring a back-illuminated sensor whose large photosensitive array is composed of 2048 x 2048 pixels (see Figure 3). By employing Princeton Instruments' exclusive XP cooling technology, these four-megapixel cameras achieve thermoelectric cooling down to -70°C via an all-metal, hermetically sealed design. This innovative cooling technology ensuresmaintenance-free operation and is backed by the industry's only lifetime vacuum guarantee.

Figure 4: PIXIS:2048 CCD cameras from Princeton
Instruments provide superb sensitivity and a large
field of view.
In addition to the XP-facilitated minimization of thermally generated (dark) noise, very high quantum efficiency and ultra-low-noise electronics make PIXIS:2048 cameras ideal for demanding, low-light-level imaging applications. Dual-speed operation (i.e., 100 kHz or 2 MHz) allows utilization in both steady-state and fast-kinetics studies.
To optimize quantitative scientific imaging performance for applications from the UV to the NIR, the PIXIS:2048 platform supports a front-illuminated CCD format, a back-illuminated format, a back-illuminated format with high UV sensitivity, and a back-illuminated format with high NIR sensitivity (note that Princeton Instruments leverages its own proprietary eXcelon® processing and back-illuminated, deep-depletion technology to deliver the highest sensitivity in the NIR while suppressing the etaloning that occurs in standard back-illuminated CCDs). Several other CCD array sizes, appropriate for various imaging and spectroscopy applications, are supported by the PIXIS series.
Complete control over all PIXIS hardware features is simple with the latest version of Princeton Instruments' 64-bit LightField® data acquisition software, available as an option. A host of novel functions for easy capture and export of imaging data are provided via the exceptionally intuitive LightField user interface
References
- https://dinnenylab.wordpress.com/glo-roots/ [accessed online in May 2016]
- R. Rellán-Álvarez et al. 2015. "GLO-Roots: an imaging platform enabling multidimensional characterization of soil-grown root systems." eLife 4 (1): e07597. doi:10.7554/eLife.07597. http://dx.doi.org/10.7554/eLife.07597.

